Lab on two-way fixed effects
08 November, 2018
The goal of the following homework is to develop our understanding of the two-way fixed effect models. See the original paper by Abowd Kramartz and Margolis.
library(data.table)
library(reshape)
library(lattice)
library(gridExtra)
library(mvtnorm)
library(ggplot2)
library(futile.logger)Constructing Employer-Employee matched data
Simulating a network
One central piece is to have a network of workers and firms over time. We then start by simulating such an object. The rest of homework will focus on adding wages to this model. As we know from the lectures, a central issue of the network will be the number of movers.
We are going to model the mobility between workers and firms. Given a transition matrix we can solve for a stationary distrubtion, and then construct our panel from there.
p <- list()
p$nk = 30 # firm types
p$nl = 10 # worker types
p$alpha_sd = 1
p$psi_sd = 1
# let's draw some FE
p$psi = with(p,qnorm(1:nk/(nk+1)) * psi_sd)
p$alpha = with(p,qnorm(1:nl/(nl+1)) * alpha_sd)
# let's assume moving PR is fixed
p$lambda = 0.05
p$csort = 0.5 # sorting effect
p$cnetw = 0.2 # network effect
p$csig = 0.5 #
# lets create type specific transition matrices
# we are going to use joint normal centered on different values
# G[i,j,k] = Pr[worker i, at firm j, moves to firm k]
getG <- function(p){
G = with(p,array(0,c(nl,nk,nk)))
for (l in 1:p$nl) for (k in 1:p$nk) {
# prob of moving is highest if dnorm(0)
G[l,k,] = with(p,dnorm( psi - cnetw *psi[k] - csort * alpha[l],sd = csig ))
# normalize to get transition matrix
G[l,k,] = G[l,k,]/sum(G[l,k,])
}
return(G)
}
G <- getG(p)
getH <- function(p,G){
# we then solve for the stationary distribution over psis for each alpha value
H = with(p,array(1/nk,c(nl,nk)))
for (l in 1:p$nl) {
M = G[l,,]
for (i in 1:100) {
H[l,] = t(G[l,,]) %*% H[l,]
}
}
return(H)
}
H = getH(p,G)
Plot1=wireframe(G[1,,],aspect = c(1,1),xlab = "previous firm",ylab="next firm")
Plot2=wireframe(G[p$nl,,],aspect = c(1,1),xlab = "previous firm",ylab="next firm")
grid.arrange(Plot1, Plot2,nrow=1)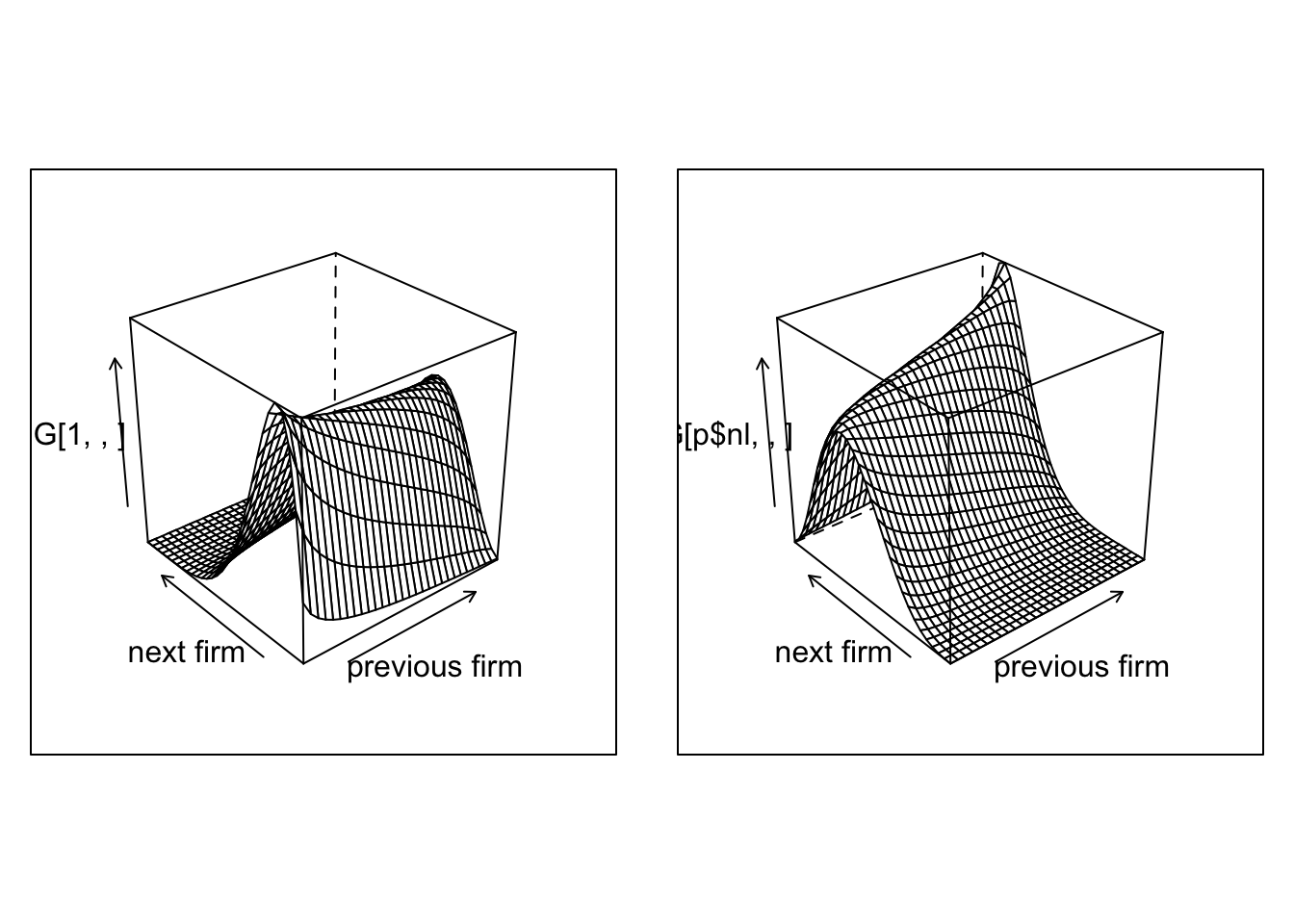
And we can plot the joint distribution of matches
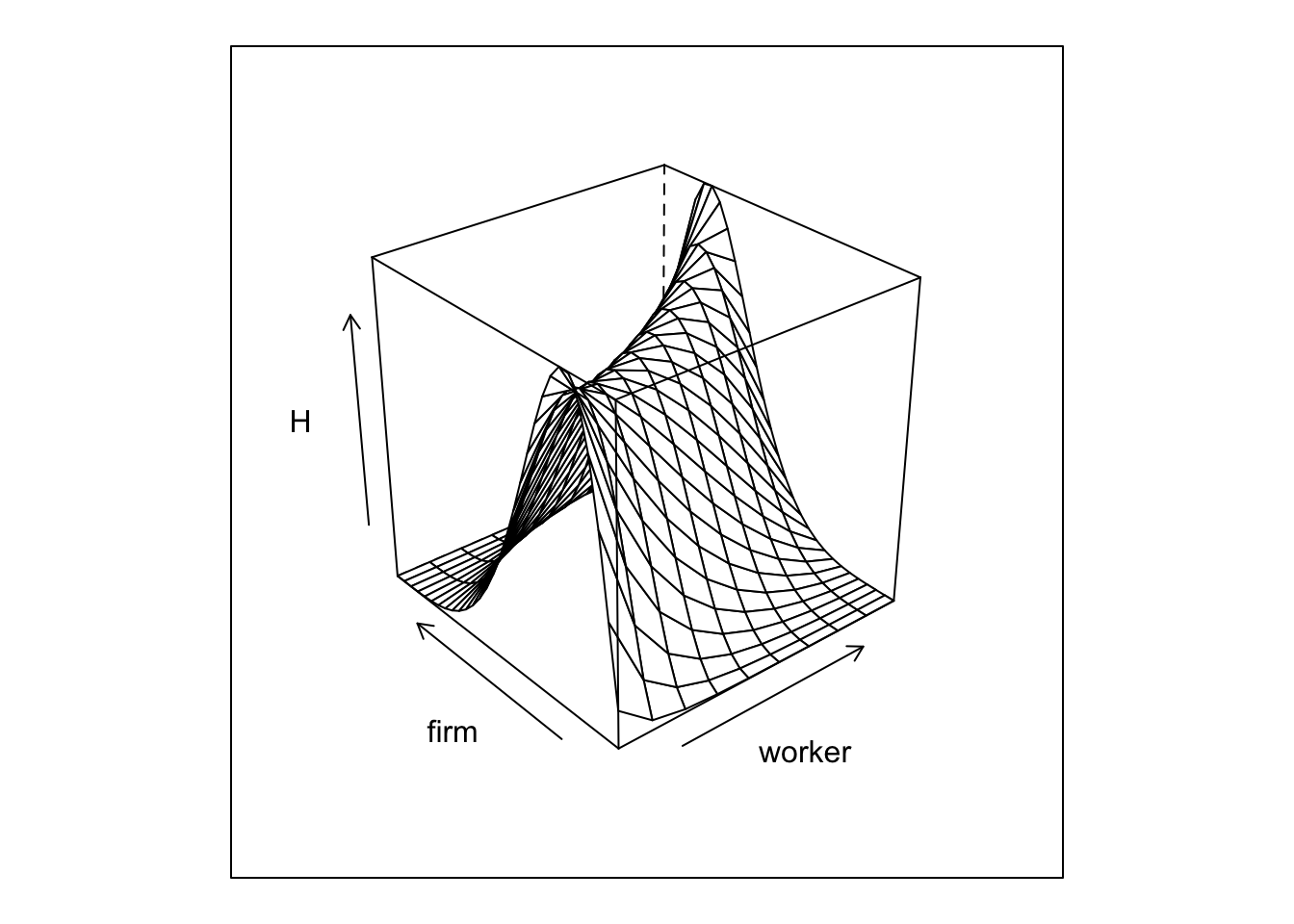
The next step is to simulate our network given our transitions rules.
p$nt = 5
p$ni = 130000
sim <- function(p,G,H){
set.seed(1)
# we simulate a panel
network = array(0,c(p$ni,p$nt))
spellcount = array(0,c(p$ni,p$nt))
A = rep(0,p$ni)
for (i in 1:p$ni) {
# we draw the worker type
l = sample.int(p$nl,1)
A[i]=l
# at time 1, we draw from H
network[i,1] = sample.int(p$nk,1,prob = H[l,])
for (t in 2:p$nt) {
if (runif(1)<p$lambda) {
network[i,t] = sample.int(p$nk,1,prob = G[l,network[i,t-1],])
spellcount[i,t] = spellcount[i,t-1] +1
} else {
network[i,t] = network[i,t-1]
spellcount[i,t] = spellcount[i,t-1]
}
}
}
data = data.table(melt(network,c('i','t')))
data2 = data.table(melt(spellcount,c('i','t')))
setnames(data,"value","k")
data[,spell := data2$value]
data[,l := A[i],i]
data[,alpha := p$alpha[l],l]
data[,psi := p$psi[k],k]
}
data <- sim(p,G,H)The final step is a to assign identities to the firm. We are going to do this is a relatively simple way, by simply randomly assigning firm ids to spells.
addSpells <- function(p,dat){
firm_size = 10
f_class_count = p$ni/(firm_size*p$nk*p$nt)
dspell <- dat[,list(len=.N),list(i,spell,k)]
dspell[,fid := sample( 1: pmax(1,sum(len)/f_class_count ) ,.N,replace=TRUE) , k]
dspell[,fid := .GRP, list(k,fid)]
setkey(dat,i,spell)
setkey(dspell,i,spell)
dat[, fid:= dspell[dat,fid]]
}
addSpells(p,data) # adds by reference to the same data.table object (no copy needed)Question 1 We are going to do some R-golfing (see wikipedia). I want you to use a one line code to evaluate the following 2 quantities:
- mean firm size, in the crossection, expect something like 15.
- mean number of movers per firm in total in our panel (a worker that moved from firm i to j is counted as mover in firm i as well as in firm j).
## [1] 17.36807## [1] 28## [1] 3.468003## [1] 62To evaluate the number of strokes that you needed to use run the following on your line of code: nchar("YOUR_CODE_IN_QUOTES_LIKE_THIS"). My scores for the previous two are 28 and 62.
Simulating AKM wages
We start with just AKM wages, which is log additive with some noise.
p$w_sigma = 0.8
addWage <- function(p,data){
data[, lw := alpha + psi + p$w_sigma * rnorm(.N) ]
}
addWage(p,data)Question 2 Before we finish with the simulation code, use this generated data to create the event study plot from Card-Heining-Kline:
- Compute the mean wage within firm
- group firms into quartiles
- select workers around a move (2 periods pre, 2 periods post)
- compute wages before/after the move for each transitions (from each quartile to each quartile)
- plot the lines associated with each transition
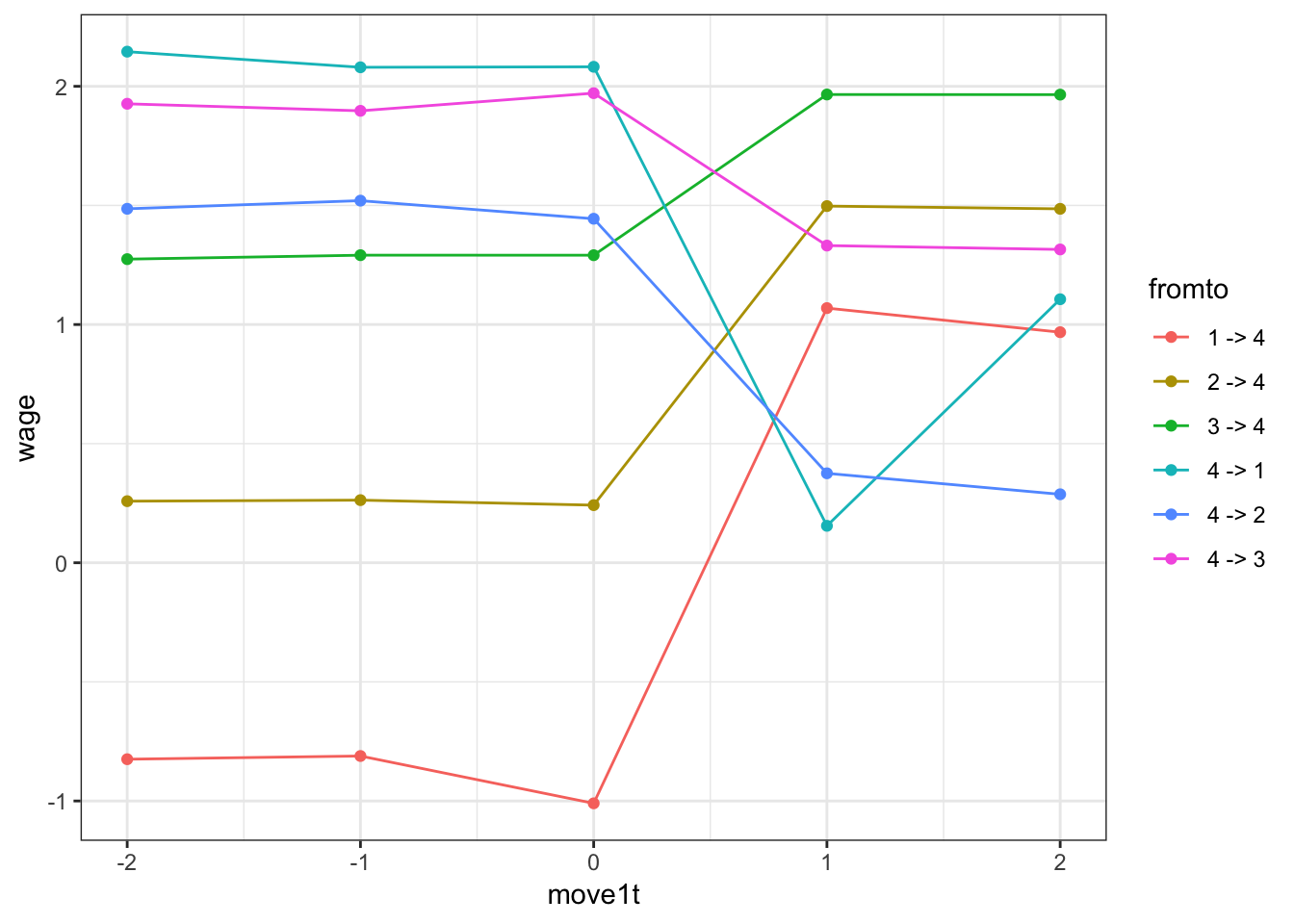
Calibrating the parameters
Question 3 Pick the parameters psi_sd,alpha_sd,csort, csig and w_sigma to roughly match the decomposition in equation (6) of Card-Heining-Kline (note that they often report numbers in standard deviations, not in variances). psi_sd, alpha_sd, w_sigma can be directly calibrated from CHK. On the other hand, csort and csig needs to be calibrated to match the numbers in CHK after AKM estimation. If AKM estimation on psi and alpha is too slow, use the true psi and alpha and get residuals directly. For this last part, however, we first need to confront the question of how to actually estimate this AKM model!
Estimating two-way fixed effects
This requires to first extract a large connected set, and then to estimate the linear problem with many dummies.
Extracting the connected set
Because we are not going to deal with extremely large data-sets, we can use off the shelf algorithms to extract the connected set. There are two approaches:
- Use the function
conCompfrom the packageggmto extract the connected set from our data. To do so you will need to construct first an adjacency matrix between the firms. An adjacency matrix is a (number offid,number offid) square matrix. Element \((i,j)=1\) if a worker ever moved from \(i\) to \(j\), else \((i,j)=0\). Here is how I would proceed to construct the adjacency matrix:- Append the lagged firm id as a new column in your data
data[ ,fid.l1 := data[J(i,t-1),fid]], for which you need to first runsetkey(data,i,t) - Extract all moves from this data set
jdata = data[fid.l1!=fid]and only keep unique pairs - Then create a column
value:=1and cast this new data to an array using theacastcommand withfill=0
- Append the lagged firm id as a new column in your data
- Use the function
compfactorfrom packagelfe. I prefer this approach because much faster.
Question 4 Use the previous procedure, extract the connected set, drop firms not in the set (I expect that all firms will be in the set).
adjMat <- function(data){
setkey(data,i,t)
data[ ,fid.l1 := data[J(i,t-1),fid]]
data <- data[complete.cases(data)]
nfirms = data[,max(fid)]
jdata = data[fid.l1!=fid][,list(fid,fid.l1)]
jdata = unique(jdata)
setkey(jdata,fid,fid.l1)
# jdata holds the indices where a sparse matrix should be 1.
amat = Matrix::sparseMatrix(i=jdata[,c(fid,fid.l1)],j=jdata[,c(fid.l1,fid)],dims=c(nfirms,nfirms))
amat
}
# amat = adjMat(data)
# g = graph.adjacency(as.matrix(amat)) # says it supports Matrix::sparsematrix, but doesn't...
# # plot(g) # takes forever, shows unconnected firms
# clu = components(g)
# connected = groups(clu)
# # keep all in largest connected set
# keep = connected$`1`
# data = data[fid %in% keep]
# or, using lfe
concomp <- function(data){
setkey(data,i,t)
data[ ,fid.l1 := data[J(i,t-1),fid]]
data <- data[complete.cases(data)]
cf = lfe::compfactor(list(f1=data[,factor(fid)],f2=data[,factor(fid.l1)]))
fr = data.frame(f1=data[,factor(fid)],f2=data[,factor(fid.l1)],cf)
data = data[fr$cf==1]
return(data)
}
data <- concomp(data)Estimating worker and firms FE
Estimating this model is non-trivial. Traditional approaches like a within-transformation is not sufficient here, as we have two fixed effects to estimate. Guimareas and Portugal propose a ZigZag estimator in the Stata Journal. Simen Gaure proposes an almost equivalent approach and develops the lfe package for R. The idea in both approaches starts with the formulation
\[ \mathbf{Y} = \mathbf{Z}\beta + \mathbf{D}\alpha + \epsilon \]
where \(\mathbf{Z}\) is \((N,k)\), and \(\mathbf{D}\) is a \((N,G_1)\) matrix of dummy variables: element \((i,j)\) of \(\mathbf{D}\) is \(1\) if \(i\) is associated to \(j\).
Both papers show the recursive relationship
\[ \left[ \begin{array}{c} \beta &=& (\mathbf{Z}'\mathbf{Z})^{-1} \mathbf{Z}'(\mathbf{Y}-\mathbf{D}\alpha) \\ \alpha &=& (\mathbf{D}'\mathbf{D})^{-1} \mathbf{D}'(\mathbf{Y}-\mathbf{Z}\beta) \end{array} \right] \]
- line 2 is just
data[,mean(resid(lm(y ~ z))),by=i] - Dimensionality of \(\mathbf{D}\) becomes irrelevant.
- Same principle for more than 1 FE:
\[ \mathbf{Y} = \mathbf{Z}\beta + \mathbf{D}_1\alpha + \mathbf{D}_2\gamma + \epsilon \]
and recursive structure:
\[ \left[ \begin{array}{c} \beta &=& (\mathbf{Z}'\mathbf{Z})^{-1} \mathbf{Z}'(\mathbf{Y}-\mathbf{D}_1\alpha-\mathbf{D}_2\gamma) \\ \alpha &=& (\mathbf{D}_1'\mathbf{D}_1)^{-1} \mathbf{D}_1'(\mathbf{Y}-\mathbf{Z}\beta-\mathbf{D}_2\gamma) \\ \gamma &=& (\mathbf{D}_2'\mathbf{D}_2)^{-1} \mathbf{D}_2'(\mathbf{Y}-\mathbf{Z}\beta-\mathbf{D}_1\alpha) \end{array} \right] \]
- First, store a
mod = data[,lm(y ~ z + D1 + D2)] - line 2 is just
data[,mean(resid(mod) + coef(mod)[3]*D2),by=i] - line 3 is just
data[,mean(resid(mod) + coef(mod)[2]*D1),by=j]
Guimaraes+Portugal do this in an iterative fashion until the difference in mean squared error (MSE) of successive estimates of line 0 becomes small.
Question 5 Write a function guimaraesPortugal that takes data and tol as an input and does the following steps:
- add 2 columns
alpha_hat=0andpsi_hat=0 - get a list of mover ids
- initiate
delta=Inf while delta>toldo- run a lin reg of
lwonpsi_hatandalpha_hat - store the
coef- if first iteration, override
coef[2:3] <- 0
- if first iteration, override
- get model residuals
- compute
MSEfor movers only - compute new
alpha_hatas mean ofres + coefs[3]*alpha_hatbyi, as above - same for
psi_hat
- run a lin reg of
- return the so updated
data.tableand check that the linear regressionlm(lw ~ alpha_hat + psi_hat)has coefficients1.00000for both FEs.
## INFO [2018-11-08 10:00:07] iter=1, MSE=0.214216, delta(MSE)=Inf
## INFO [2018-11-08 10:00:14] iter=10, MSE=0.097778, delta(MSE)=0.000759
## INFO [2018-11-08 10:00:21] iter=20, MSE=0.093468, delta(MSE)=0.000246
## INFO [2018-11-08 10:00:29] iter=30, MSE=0.091924, delta(MSE)=0.000116
## INFO [2018-11-08 10:00:36] iter=40, MSE=0.091147, delta(MSE)=0.000067
## INFO [2018-11-08 10:00:43] iter=50, MSE=0.090681, delta(MSE)=0.000044
## INFO [2018-11-08 10:00:50] iter=60, MSE=0.090371, delta(MSE)=0.000030
## INFO [2018-11-08 10:00:57] iter=70, MSE=0.090155, delta(MSE)=0.000022
## INFO [2018-11-08 10:01:03] iter=80, MSE=0.089996, delta(MSE)=0.000016
## INFO [2018-11-08 10:01:11] iter=90, MSE=0.089877, delta(MSE)=0.000013
## INFO [2018-11-08 10:01:20] iter=100, MSE=0.089786, delta(MSE)=0.000010
## INFO [2018-11-08 10:01:27] iter=110, MSE=0.089714, delta(MSE)=0.000008
## INFO [2018-11-08 10:01:34] iter=120, MSE=0.089656, delta(MSE)=0.000006
## INFO [2018-11-08 10:01:41] iter=130, MSE=0.089610, delta(MSE)=0.000005
## INFO [2018-11-08 10:01:49] iter=140, MSE=0.089572, delta(MSE)=0.000004
## INFO [2018-11-08 10:01:57] iter=150, MSE=0.089541, delta(MSE)=0.000004
## INFO [2018-11-08 10:02:05] iter=160, MSE=0.089515, delta(MSE)=0.000003
## INFO [2018-11-08 10:02:12] iter=170, MSE=0.089493, delta(MSE)=0.000003
## INFO [2018-11-08 10:02:19] iter=180, MSE=0.089475, delta(MSE)=0.000002
## INFO [2018-11-08 10:02:26] iter=190, MSE=0.089459, delta(MSE)=0.000002##
## Call:
## lm(formula = lw ~ alpha_hat + psi_hat, data = data)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.70312 -0.21353 -0.00044 0.21438 1.53969
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.0002860 0.0004406 0.649 0.516
## alpha_hat 1.0001969 0.0013660 732.223 <2e-16 ***
## psi_hat 1.0004850 0.0019395 515.842 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.3175 on 519317 degrees of freedom
## Multiple R-squared: 0.5281, Adjusted R-squared: 0.5281
## F-statistic: 2.905e+05 on 2 and 519317 DF, p-value: < 2.2e-16Note You can increase speed by focusing on movers only first to recover the psi.
Question 5 Now do the same thing but use the function felm from package lfe. Write function gaureAKM which takes data and p as inputs.
## INFO [2018-11-08 10:02:27] running gaureAKM with lambda=0.050000
## INFO [2018-11-08 10:02:41] done.Limited mobility bias
We now have every thing we need to look at the impact of limited mobility bias. Compute the following:
- Compute the estimated variance of firm FE
- Do it for varying level of mobility \(\lambda\). Collect for each the number of movers, the actual variance and the estimated variance. Run it for diffenrent panel lengths: 5,6,8,10,15.
Question 6 Report this in a plot. Fix T and vary lambda. Plot (i) correlation between firm fixed effect and individual fixed effect and (ii) variance of firm fixed effect against the number of movers. This should look like the Andrews et al. plot.
p$nlambda = 10
pdat = data.frame(lambda=seq(from=0.03,0.5,length.out = p$nlambda),varFid = 0, cov_alpha_psi = 0,varFid_true = 0, cov_alpha_psi_true = 0)
for (i in 1:nrow(pdat)){
p$lambda = pdat[i,"lambda"]
flog.info("---> doing for lambda = %f",p$lambda)
data = buildData(p)
data = concomp(data)
g <- gaureAKM(p,data)
pdat[i,"varFid"] <- var(g$psi$effect)
pdat[i,"cov_alpha_psi"] <- g$data[!is.na(psi_hat),cov(psi_hat,alpha_hat)]
pdat[i,"varFid_true"] <- g$data[,var(psi)]
pdat[i,"cov_alpha_psi_true"] <- g$data[,cov(psi,alpha)]
}## INFO [2018-11-08 10:02:41] ---> doing for lambda = 0.030000
## INFO [2018-11-08 10:02:46] running gaureAKM with lambda=0.030000
## INFO [2018-11-08 10:03:24] done.
## INFO [2018-11-08 10:03:24] ---> doing for lambda = 0.082222
## INFO [2018-11-08 10:03:29] running gaureAKM with lambda=0.082222
## INFO [2018-11-08 10:03:37] done.
## INFO [2018-11-08 10:03:37] ---> doing for lambda = 0.134444
## INFO [2018-11-08 10:03:42] running gaureAKM with lambda=0.134444
## INFO [2018-11-08 10:03:47] done.
## INFO [2018-11-08 10:03:47] ---> doing for lambda = 0.186667
## INFO [2018-11-08 10:03:52] running gaureAKM with lambda=0.186667
## INFO [2018-11-08 10:03:56] done.
## INFO [2018-11-08 10:03:56] ---> doing for lambda = 0.238889
## INFO [2018-11-08 10:04:02] running gaureAKM with lambda=0.238889
## INFO [2018-11-08 10:04:06] done.
## INFO [2018-11-08 10:04:06] ---> doing for lambda = 0.291111
## INFO [2018-11-08 10:04:13] running gaureAKM with lambda=0.291111
## INFO [2018-11-08 10:04:17] done.
## INFO [2018-11-08 10:04:17] ---> doing for lambda = 0.343333
## INFO [2018-11-08 10:04:24] running gaureAKM with lambda=0.343333
## INFO [2018-11-08 10:04:27] done.
## INFO [2018-11-08 10:04:27] ---> doing for lambda = 0.395556
## INFO [2018-11-08 10:04:33] running gaureAKM with lambda=0.395556
## INFO [2018-11-08 10:04:36] done.
## INFO [2018-11-08 10:04:37] ---> doing for lambda = 0.447778
## INFO [2018-11-08 10:04:44] running gaureAKM with lambda=0.447778
## INFO [2018-11-08 10:04:47] done.
## INFO [2018-11-08 10:04:47] ---> doing for lambda = 0.500000
## INFO [2018-11-08 10:04:54] running gaureAKM with lambda=0.500000
## INFO [2018-11-08 10:04:57] done.p1=ggplot(data=pdat,mapping=aes(x=lambda,y=varFid)) + geom_line() + ggtitle("var(psi)")
p2=ggplot(data=pdat,mapping=aes(x=lambda,y=cov_alpha_psi)) + geom_line()+ ggtitle("cov(alpha,psi)")
grid.arrange(p1, p2,nrow=1)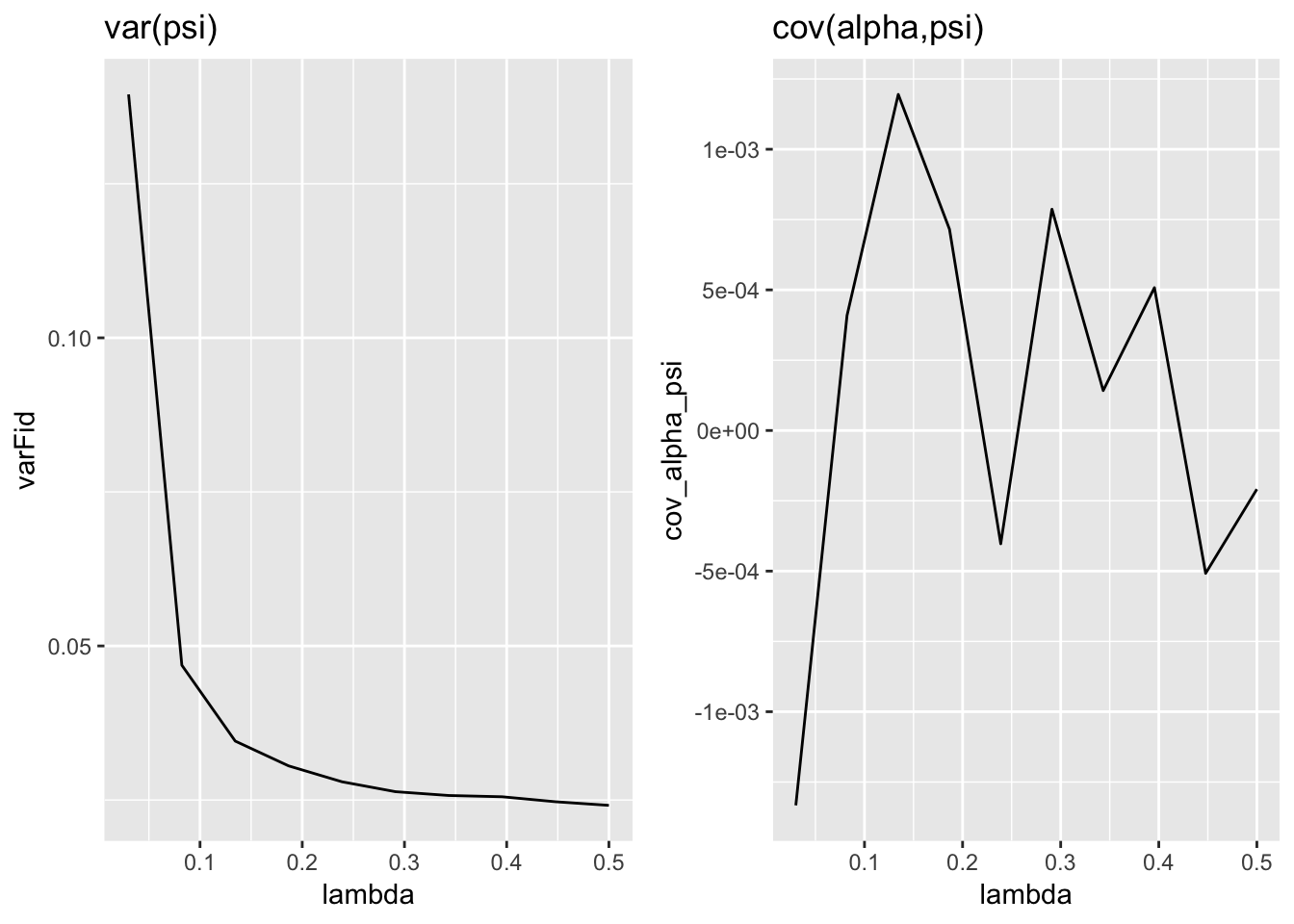
Alleviating the bias using Split Sample Jackknife
Pick a relatively short \(T\) together with a low \(\lambda\). Simulate one data-set. Next conduct the following procedure:
- Estimate AKM on the full sample
- Split your sample within firm, ie, within firm, split your movers in a balanced way between group 1 and group 2. Do the same for the individuals who don’t move. You can do that by assigning a random number to each worker within firm and then defining the group as being below or above the median.
- Perform AKM on eash split-sample
- Form average estimates of each parameter by just averaging over the subpanels: \(\overline{\theta}_{1/2} = 0.5 \hat{\theta}_{1} + 0.5 \hat{\theta}_{2}\)
- Compute biased-corrected estimates for variance of firm-effects and covariances between worker and firm effects using the Split Panel Jackknife by Dhaene and Jochmans: use \(2 \hat{\theta} - \overline{\theta}_{1/2}\)
The theta in the bias correction formula is on var(psi), not psi itself.
Question 7 Report the true values, the non biased corrected and the bias corrected.